A recent paper in PLoS Biology by David Goldstein’s group is being seen as another ‘death of GWAS’ moment (again?). I have a lot of issues with this paper, but I will be brief and stick to my main objection; the authors attempt to demonstrate that common associations can be caused by sets of rare variants, and in doing so inadvertantly show they most of them are not.
The Paper and the Press
This is another example of a scientific paper being careful to make no solid, controversial claims, but being surrounded by a media story that is not justified by the paper itself. The only real solid claim in the paper is that, if you do not include rare SNPs in your genome-wide association study, and rare SNPs of large effect are contributing to disease, then you will sometimes pick up more common SNPs as associated, because they are in Linkage Disequilibrium with the rare SNPs. Pretty uncontroversial, in so far as it goes. The paper makes no attempt to say whether this IS happening, just says that it CAN happen, and that we should be AWARE of it.
However, in the various articles around the internet, this paper is being received as if it makes some fundamental claim about complex disease genetics; that this somehow undermines Genome-Wide Association Studies, or shows their results to be spurious. David Goldstein is quoted on Nature News:
…many of the associations made so far don’t seem to have an explanation. Synthetic associations could be one factor at play. Goldstein speculates that, “a lot, and possibly the majority [of these unexplained associations], are due to, or at least contributed to, by this effect”.
Another author is quoted here as saying
We believe our analysis will encourage genetics researchers to reinterpret findings from genome-wide association studies
Much of the coverage conflates this paper with the claim that rare variants may explain ‘missing heritability’, which is an entirely different question; Nature News opens with the headline “Hiding place for missing heritability uncovered”. Other coverage can be found on Science Daily, Gene Expression and GenomeWeb.
Does this actually happen?
Is all this fuss justified? How common is this ‘synthetic hit’ effect; are a lot of GWAS hits caused by it, or hardly any? There are many ways that you could test this; for instance, you could make some predictions about what distribution of risk you’d expect to see in the many fine mapping experiments that have been done as follow ups to Genome-Wide Association Studies (this would be trivially easy to do using the paper’s simulations).
However, there is an even easier way to test the prevalence of the effect. If most GWAS hits are tagging relatively common variants, then you would expect to see most disease associated SNPs with a frequency in the 10% to 90% range (the range for which GWAS are best powered). However, a SNP with a frequency of 50% is less likely than one with a frequency of 10% to tag a SNP with frequency 0.5%, so if most GWAS hits are tagging rare variants, then you would expect to see most associated SNPs with a frequency skewed towards the very rare or the very common.
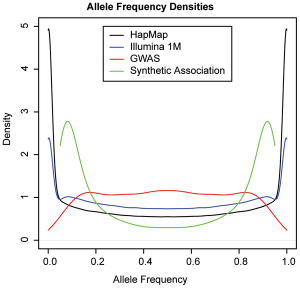
In fact, the paper makes an explicit calculation of the expected frequency distribution of GWAS hits, under their synthetic model. In-double-fact, the paper plots this distribution against the distribution of know GWAS hits. And here is that plot, taken directly from the paper (Figure 5):
The green line is the expected frequency distribution of ‘synthetic’ associations; the red line is the actual distribution. We can see that the GWAS hits we do see fail to follow the distribution for synthetic associations; in fact, they follow pretty much exactly the distribution we’d expect if most common associations are tagging common causal SNPs.
The paper manages to pretty conclusively show both that demonstrate that synthetic SNPs can occur, but they rarely do.
Dickson, S., Wang, K., Krantz, I., Hakonarson, H., & Goldstein, D. (2010). Rare Variants Create Synthetic Genome-Wide Associations PLoS Biology, 8 (1) DOI: 10.1371/journal.pbio.1000294